In type 2 diabetes (T2D), pancreatic islet beta cells gradually become unable to secrete enough insulin to regulate blood glucose levels. But what are the molecular processes taking place inside these cells along the road to diabetes? Insights into such processes would have a major impact on diabetes prevention and treatment but have largely eluded scientists thus far. Thanks to a unique dataset from living donors, and innovative multi-omics analysis, an international research team led by investigators at the Paul-Langerhans-Institute Dresden (PLID), the SIB Swiss Institute of Bioinformatics and the Max-Planck-Institute (MPI) of Biochemistry has shed some light into the processes driving islet dysfunction. The results of this collaborative study, part of the IMI RHAPSODY project, are published in the journal Nature Metabolism.
About RHAPSODY
This study was funded in part by the Innovative Medicines Initiative RHAPSODY, a public-private partnership between the European Union – represented by the European Commission – and the pharma industry, which unites researchers and experts from 26 partner institutions in both the public and private sectors, and where SIB is offering its expertise to coordinate the integration and analysis of clinical data.
Not all T2D beta cells become dysfunctional in the same way
In people with T2D, islet beta cells have been hypothesized to become dysfunctional following a ‘de-programming’, leading them back to their precursor state. However, this study shows for the first time that the gene expression and proteomic profile of islet cells in individuals with T2D is very divergent, whereas it is remarkably homogeneous in islet cells of non-diabetic subjects. In other words, beta cells appear to become dysfunctional by different molecular processes in different people, rather than follow the same ‘reset’ pathway in all individuals. This reinforces the idea that there are no single one-size fits all way to treat T2D and supports the need for a personalized approach.
Potential biomarkers to monitor disease
Thanks to innovative integration of lipidomics, transcriptomics and clinical data from living donors (read below), performed by SIB’s Computational biology group Vital-IT, the study identified several pancreatic islet genes, and lipids retrieved from blood plasma of the donors, associated with high blood sugar.
- ALDOB (Aldolase B) stands out as being the gene most associated with hyperglycemia and its expression was found to be strongly elevated in T2D at both transcript and protein levels.
- In addition, the study revealed for the first time a set of genes whose expression was already altered in patients with impaired glucose tolerance, but who were not yet diagnosed with T2D.
- Finally, several lipid classes (i.e. phospholipids and dihydroceramides) were also found as potential biomarkers for disease.
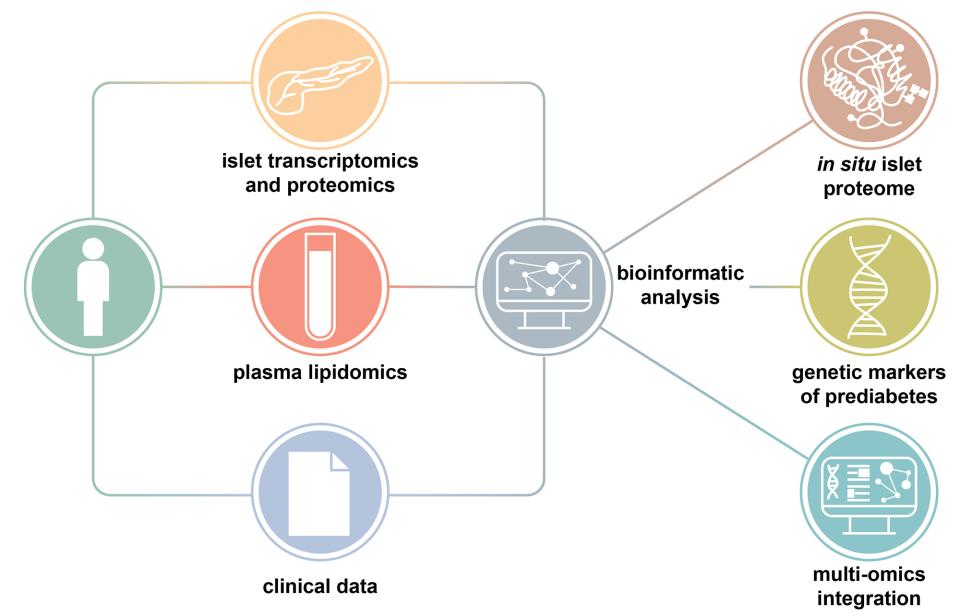
A valuable resource to investigate the prediabetic-diabetic continuum
So far, the subtle molecular processes taking place along the road to diabetes eluded scientists because human pancreatic islet data usually came from deceased donors. “In contrast, this study provides the first extensive analysis on islets in situ and plasma samples from the largest cohort of in-depth metabolically profiled living donors”, says Leonore Wigger, Computational Scientist at SIB and co-first author on the study. This approach allowed to circumvent the usual shortcomings associated with data from deceased donors, which can only be characterized as diabetic or non-diabetic, but also are in a particular state of metabolic stress associated with the donor’s terminal condition, and lacking extensive clinical and laboratory information. “Multi-omics data were thus generated using state-of-the-art approaches and integrated in a fashion not previously used in studies on islet dysregulation in relation to hyperglycemia in humans.”
Mark Ibberson, Director of SIB's Computational biology group Vital-IT and co-Principal investigator of the study concludes: “The results of this study change the way that we think about how the pancreas gets damaged in T2D, and provide a unique dataset of transcriptomic and proteomic data that can be used to enable further discoveries, for instance as an addition to existing datasets.”
Reference(s)
Wigger L, Barovic M, Brunner A-D et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories toward type 2 diabetes. Nature Metabolism 2021.