An improved view of influenza evolution with CoalRe

“Accounting for reassortment events reduces bias in influenza inference”. In this in silico talk, SIB’s Ugnė Stolz from Tanja Stadler’s Group at ETH Zurich introduces CoalRe, a framework enabling to take into account reassortment when inferring virus evolution and infection pathways. In a recent paper published in PNAS, the team showed that this framework permits better estimates of effective population size and evolutionary rates. CoalRe is freely available as an add-on to the BEAST2 phylogenetic software.
About the in silico talks series – The latest in bioinformatics by SIB Scientists
The in silico talks online series aims to inform bioinformaticians, life scientists and clinicians about the latest advances led by SIB Scientists on a wide range of topics in bioinformatics methods, research and resources. Stay abreast of the latest developments, get exclusive insights into recent papers, and discover how these advances might help you in your work or research, by subscribing to the in silico talks mailing list.
This work was featured as part of the latest SIB Remarkable Outputs 2019: a list of ‘must-read’ works by SIB Members produced in 2019 and selected by a committee of SIB Group Leaders and Researchers
The pathway of a viral infection is usually inferred from a ‘snapshot’ of the current viral population by sampling infected individuals. This enables the reconstruction of the virus’ phylogenetic tree, which offers a way to look back in time at the history of an epidemic.
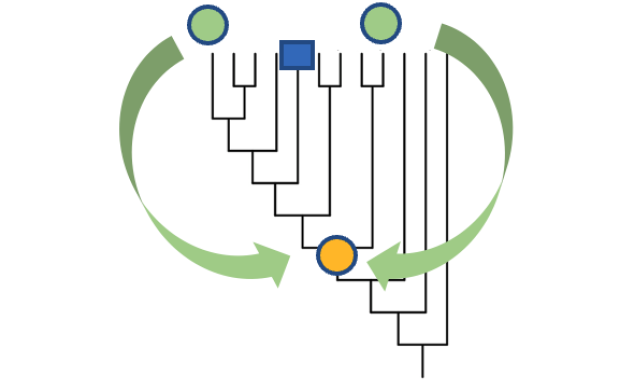
However, this inference approach is made much more difficult for specific viruses, such as influenza, whose genome is made up of several distinct RNA molecules. Such ‘segmented viruses’ have the property of exchanging the different parts of their parental genome as they replicate within hosts (reassortment events), leading to a hybrid progeny. A person infected by two different strains of the virus can thus exhibit novel variant strains.
For such viruses, infection pathways can no longer be defined by trees, but by networks, to include the cross-over aspect of reassortment. New evolutionary models are needed for this, and CoalRe is one such framework.
Listen to Ugnė as she presents the model, demonstrates its use for several influenza A and B strains, and guides you through a tutorial to understand how to apply and run CoalRe for your datasets.