For the first time, biodata from humans have been integrated with that of other organisms to provide the most comprehensive picture of human gene function to date. The new ‘PAN-GO’ resource used evolutionary modelling co-developed by SIB scientists to assign known functions to over 20,000 human genes. The work involved more than 150 biologists contributing to the international Gene Ontology Consortium, and is published today in the journal Nature.
Innovative bioinformatics creates a new biomedical resource
Researchers carrying out big data biomedical studies can now gain more accurate and informative insights into human disease, cell biology and more. The PAN-GO functionome resource overcomes gaps in experimental data on human gene functions, by integrating data from related genes in model organisms – including mice, zebrafish, fruit flies, yeast, and even plants. The new resource is openly available to everyone, and structured in a machine-readable format that enables artificial intelligence and other computational data analyses.
PAN-GO is part of the Gene Ontology (GO) knowledgebase developed by the Gene Ontology Consortium, which works to provide comprehensive, up-to-date information on gene function across the tree of life. Funded by NIH, GO is cited by over 30,000 publications each year for its use in the analysis and interpretation of biodata. Scientists in SIB’s Swiss-Prot group are members of the consortium and have contributed to GO since its inception 25 years ago.
Developed by SIB’s Swiss-Prot group, the Keck School of Medicine of the University of Southern California (USC), and other institutions, the new resource combines expertise in extracting species-specific information on protein function from the scientific literature and generalizing this information across species through cutting-edge, large-scale evolutionary modelling. Thanks to this innovative approach, known functions are now assigned to over 20,000, or 82%, of human protein-coding genes.
PAN-GO contributes to Swiss-Prot’s work to generate machine-readable knowledge of biology and complements UniProt, the leading protein knowledgebase co-developed by the group. Its development aligns with SIB’s mission to push the boundaries of data science, accelerate innovation across medicine and biodiversity, and ensure that biological knowledge is widely accessible for the benefit of science and society.
PAN-GO is part of the Gene Ontology (GO) knowledgebase developed by the Gene Ontology Consortium, which works to provide comprehensive, up-to-date information on gene function across the tree of life. Funded by NIH, GO is cited by over 30,000 publications each year for its use in the analysis and interpretation of biodata. Scientists in SIB’s Swiss-Prot group are members of the consortium and have contributed to GO since its inception 25 years ago.
A wealth of new information from evolutionary modelling
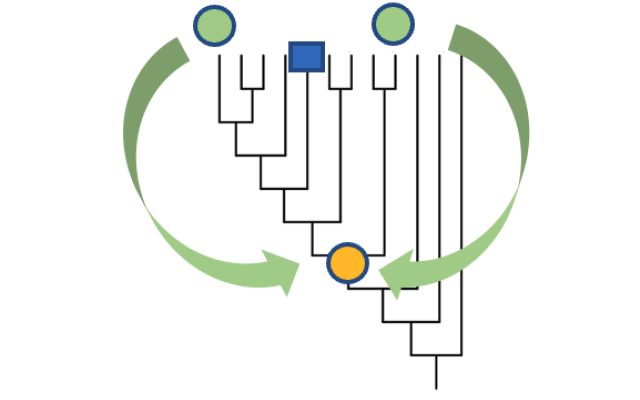
Gene function has traditionally been determined at the level of a specific gene in a specific species, and in one of two ways: experimental data and computational predictions. PAN-GO’s evolutionary modelling provides a powerful third method – identifying over twice as many functional characteristics for human genes than currently available through curated experimental data on human genes, and around three times as many than predicted by computational tools.
New biological insights and experimental directions
PAN-GO’s developers showed the resource generates clearer and more informative insights than previously available from computational genomics analyses – such as when comparing genes expressed in a specific type of cancer cell to the corresponding normal cell type.
The evolutionary models themselves can be used to examine how and when different gene functions arose. An initial analysis shows that most human genes have performed the same function for hundreds of millions of years or more, even before our ancestors were animals.
PAN-GO will also help guide future research on the roughly 3,600 human protein-coding genes whose biological function remains unknown, as well as the thousands more whose functions are only partially known. Researchers can submit suggestions for updating the resource through its website, helping to ensure continued improvements over time.
PAN-GO and UniProt: complementary and mutually beneficial
The evolutionary models used to create PAN-GO were built using reference protein sets (proteomes) in UniProt for different species. The models also harnessed functional annotations in UniProt and other databases created by GO Consortium members – that is, experimental evidence on protein function identified in the scientific literature by expert biocurators and provided with the corresponding protein sequence in the databases.
Gene entries on PAN-GO and the broader GO knowledgebase link to the corresponding protein entries on UniProt, and functional annotations in PAN-GO are imported into UniProt. The new annotations also enable SIB’s biocurators to search the literature for experimental data to confirm these evolutionarily inferred functions. This complementary relationship enhances UniProt’s value as a highly reliable source of the latest scientific knowledge on proteins.
Reference(s)
Feuermann, M., Mi, H., Gaudet, P. et al. A compendium of human gene functions derived from evolutionary modelling. Nature (2025).
Image: Adapted from Extended Data Figure 1 from the article