From where did the new coronavirus (nCoV, see box) arise, and how did it move to humans? How is it spreading and evolving? How can we develop therapies to treat it? SIB Groups provide a range of tools and resources that can help researchers answer these questions. These tools link viral epidemiology and genomics to protein sequences, variants, functions, and three-dimensional structures. Find out more about some of these tools below.
Coronavirus: some biological facts
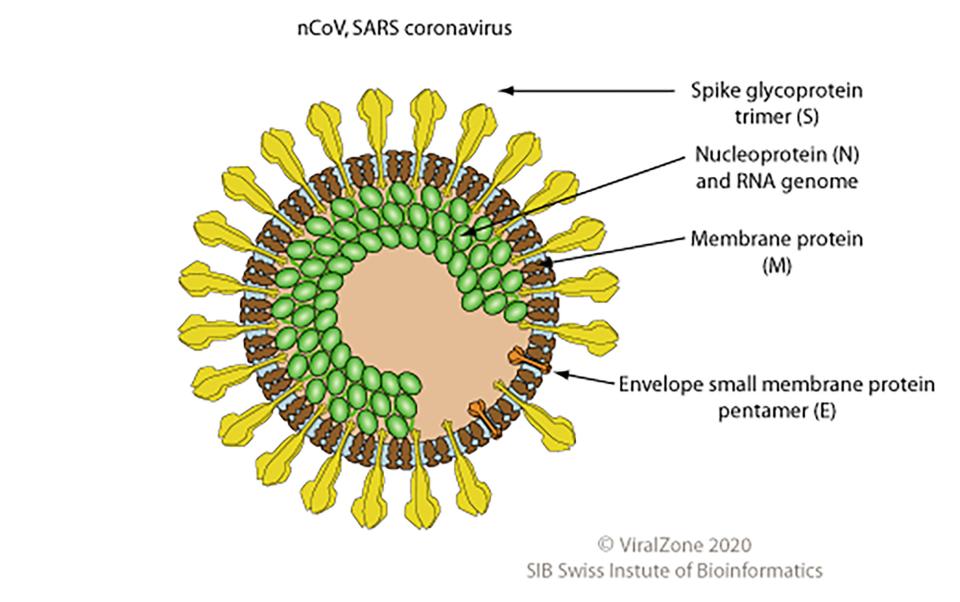
The 2019–20 coronavirus, first identified in Wuhan, the capital of China's Hubei province, is an enveloped single-stranded RNA virus. The name ‘Coronavirus’ stems from the Latin corona (i.e. crown), and refers very aptly to the shape of the proteins around the virion (see image). Coronaviruses are found in many vertebrate species and cause respiratory diseases, such as the common cold or SARS. The more recent nCoV coronavirus has emerged from a still unknown animal reservoir and can be transmitted from human to human.
Getting to know the coronavirus: from genomics and epidemiology…
The first coronavirus genome sequence was published on 10 January 2020. Since then, the NextStrain resource, co-developed by the SIB Group of Richard Neher (University of Basel), incorporates nCoV genomes as soon as they are shared publicly, and enables clinical researchers to track in near-real time how the coronavirus genome evolves – information that will be crucial in the efforts to combat the spread of the disease.
…to knowledge of viral proteins, their functions and structures…
The coronavirus genome encodes 13 (or possibly 14) proteins. These viral proteins control the virus’ transcription, replication, and spread throughout the host population.
Knowledge of viral protein sequences and how they function is now available as a pre-release of the coronavirus entries of the UniProt universal protein knowledgebase – an international collaboration between SIB's Swiss-Prot Group, led by Alan Bridge, EMBL-EBI, and PIR.
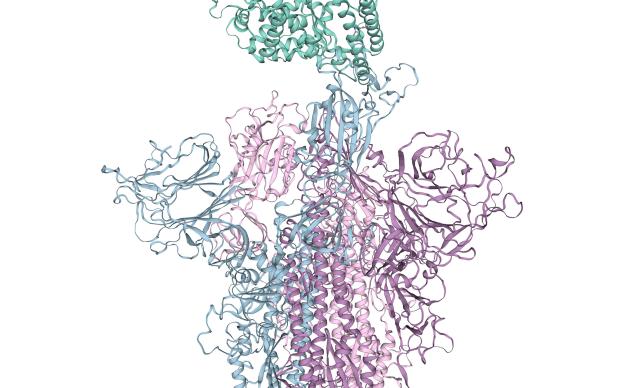
Combined with data on viral genome evolution and epidemiology, knowledge of viral proteins will be crucial to aid clinical researchers in their efforts to design new therapies. SWISS-MODEL, developed by the SIB Group of Torsten Schwede at the University of Basel, can for instance generate three-dimensional models of those viral proteins (see dedicated page) – models which shed further light on their evolution and functional properties (as in these two recent examples here and here) – and, potentially, their weaknesses, in the context of drug development.
A new nCoV-dedicated page on ViralZone provides further biological insights, including a detailed comparison with the SARS virus’ genome as well as cross-links to complementary resources mentioned above. “We are highlighting specific differences between nCoV and SARS, in particular in its gene composition that seems closer to bat viruses and may explain the difference in pathogenicity and transmission”, explains Philippe Le Mercier who leads the developments of ViralZone at SIB’s Swiss-Prot Group.