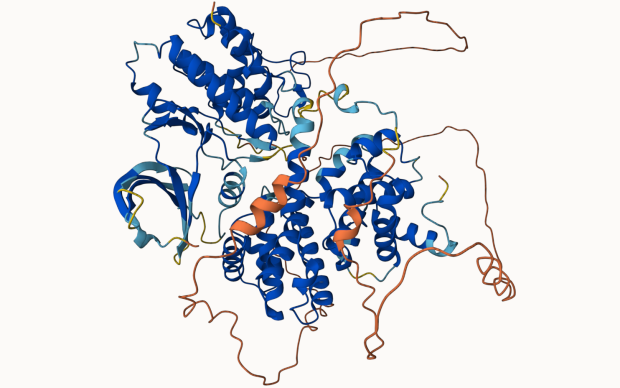
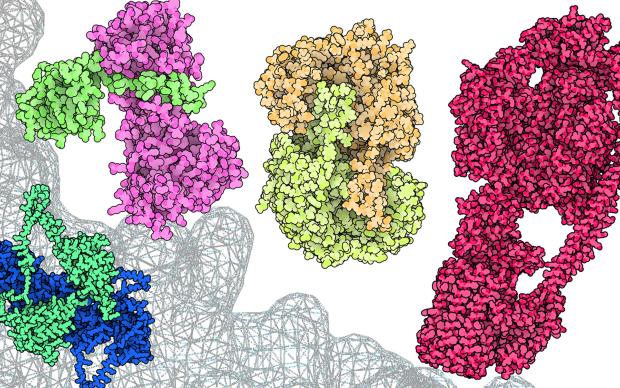
In the Computational Structural Biology (CSB) Group, we focus on the development of methods and algorithms to model, simulate and analyse three-dimensional protein structures and their molecular properties in order to apply these techniques to the understanding of biological processes at a molecular level. Our main emphasis is on homology modelling – using evolutionary information to model protein tertiary and quaternary structures. Applications in biomedical research include the study of protein-ligand interactions, drug discovery, structure-guided protein engineering, and the interpretation of mutations related to diseases and drug resistance.
The group develops the SIB Resource SWISS-MODEL, a protein structure homology-modelling suite of software tools and databases.