ModelArchive, an open repository for sharing computationally determined protein structure models, has been awarded a grant by swissuniversities to expand open research data (ORD) principles. Several SIB group leaders are collaborating with international experts to develop this data resource, which complements the Protein Data Bank (PDB) for experimentally determined protein structures.
Harnessing the explosion of predicted protein structures
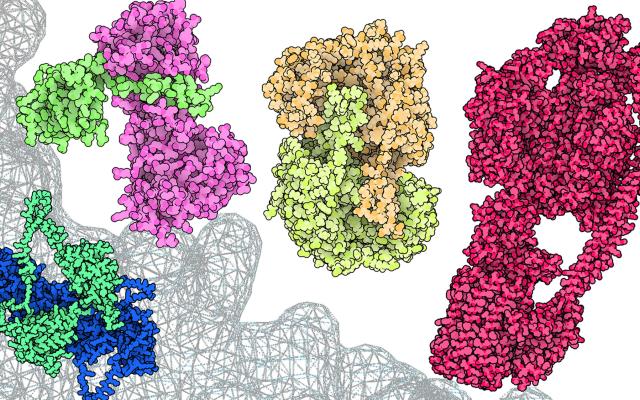
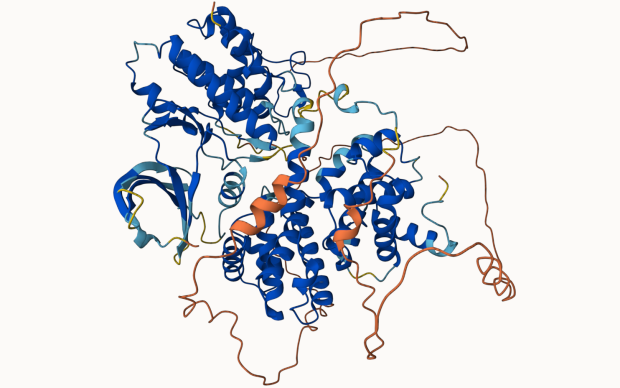
Novel modelling methods powered by deep learning, such as AlphaFold, now allow prediction of protein structures at a high quality, reaching near-experimental accuracy. This has resulted in an explosion of computer-predicted models, which complement the otherwise limited range of experimentally determined structures. Furthermore, advances are already being observed in the modelling of protein complexes such as of their interactions with small molecules (e.g. drugs), RNA and DNA. The modelling of proteins in different conformational states is also progressing as well as in protein engineering and design studies. All these models need a dedicated space to be stored and made available to life scientists for use in their respective fields of research, from drug discovery to molecular biology.
This is where ModelArchive comes in. Developed in the group of SIB's Torsten Schwede, it enables protein structures determined by computational methods to be deposited and shared. It thus complements the Protein Data Bank (PDB) and PDB-Dev which store structures derived and partially derived from experimental data respectively. By building and expanding on current open research data best practices such as the ModelCIF data format, ModelArchive aims to become the reference repository for sharing computationally modelled protein structures. This initiative to foster open research data (ORD) has been awarded funding by swissuniversities (See box).
The call from swissuniversities for projects fostering open research data
In 2022, Swissuniversities launched a call for projects which support and promote excellence in Open Research Data practices. This is part of the Swiss National Open Research Data Strategy’s Open Science I Phase B – ORD programme which it was commissioned to develop by the State Secretariat for Education, Research and Innovation (SERI). Several projects involving or led by SIB members have been granted funding, evidence of SIB’s role in catalyzing ORD.
A treasure trove of data to foster new discoveries
The initiative is led by several scientists with a long-standing expertise in computational structural biology namely Andrea Cavalli, Matteo Dal Peraro, Markus Lill, Olivier Michielin, Torsten Schwede and Vincent Zoete, all SIB Group Leaders. On this project being awarded funding, Torsten Schwede says, “more than 74,000 models are currently available in ModelArchive with this number set to increase rapidly in the future. With this support from swissuniversities, we can advance the current ORD best practices and increase the reusability of computed structure models. Combining experimental structures with computational models provides structural biologists with a treasure trove of data to foster new discoveries.”
Building upon and establishing new open research data best practices
Making protein structure data freely available is at the heart of the efforts of the team that develops and maintains ModelArchive. Expanding on these ORD practices will not only involve ensuring that sharing of data is implemented in a FAIR (Findable, Accessible, Interoperable and Reusable) manner but also that the values of these practices are shared with other stakeholders such as journals, funding agencies and life science researchers. As stated in their proposal, these practices align with the SwissBioData ecosystem (SBDe) co-led by SIB, which aims to significantly advance FAIR data sharing in Switzerland.